SWITCHES model structure
In the SWITCHES database, reaction networks (topologies) are classified according to the number of reactants (M) and the number of reactions (N) in the topology, in the format MxN. So a topology belonging to the class 3x3 has 3 molecules and 3 reactions. A topology that has been assigned reaction parameters (reaction rates and concentrations) is called a model.
We describe the file format using an example model with 3 molecules and 3 reactions.
| M3x3.111.151: | |AabX|DabX|Jacb| | 0.0314 | 0.6423 | 0.0231 | 0.0368 | 0.2076 | 1.5814 | 1.3272 | 0.4259 | 0.0559 |
Here, the string following M and before ':' (M3x3.111.151) uniquely identifies all topologies and all models using the format class_id.topology_id.model_number i.e (3x3 class_id, 111 topology, 151 model_number). The section after the ':' and before the numbers begin is the model signature (|AabX|DabX|Jacb|).
This specifies the configuration of reactions in the topology, delimited by vertical bars '|'. The first letter of each block (delimited by '|') in the signature specifies the reaction from the list below, and the following lower case letters identify the molecules involved in that reaction. X indicates that the reaction only involves two molecules so there is no third entry. Within this format, the reactions are ordered aphabetically.
The reactions are taken from this table of basic reaction types.The first letter (upper caps) describes the reaction type (A-L). The next three letters (lower caps) describe the reactant labels.
| a <==> b | AabX | Conversion reaction |
| 2a <==> b | BabX | Conversion reaction |
| a --a--> b | CabX | Enzymatic reaction catalyzed by a |
| a --b--> b | DabX | Enzymatic reaction catalyzed by b |
| a <==> b + c | Eabc | Conversion reaction |
| 2a <==> b + c | Fabc | Conversion reaction |
| 2a + b <==> c | Gabc | Conversion reaction |
| 2a + b <==> 2c | Habc | Conversion reaction |
| 4a + b <==> c | Iabc | Conversion reaction |
| a --b--> c | Jabc | Enzymatic reaction catalyzed by b |
| a --a--> b + c | Kabc | Enzymatic reaction catalyzed by a |
| a --b--> b + c | Labc | Enzymatic reaction catalyzed by b |
All enzymatic reactions are shortcuts for a full mass-action implementation of the following two reactions
E + S <======> E.S -----> E + P
E = Enzyme
S = Substrate
P = Product.
We explicitly compute the concentration of the E.S complex, and the two reactions involving it, for every enzyme.
Following the signature is a list of values that specify the parameters of the model, in the following sequence:
First, all the molecule concentrations, in alphabetical order.
Then, each of the reaction parameters, in alphabetical order. Each reaction requires two parameters, as follows:
Reversible, non-enzymatic reactions :
- Parameter 1 = Kf
- Parameter 2 = Kb
Enzymatic reactions :
- Parameter 1 = kcat
- Parameter 2 = Km
Reactions are converted from the Michaelis-Menten into the mass-action form while simulating in MOOSE. Here, enzymatic activity is described by three parameters.
These are:
| k1 | | k3
| |
| E + S | <======> | E.S | -------> | E + P |
| k2 | | | |
k1 = (k2 + k3 )/Km
k2 = 4 * kcat
k3 = kcat
---------------------------------------------------------------------------------------------------------------------
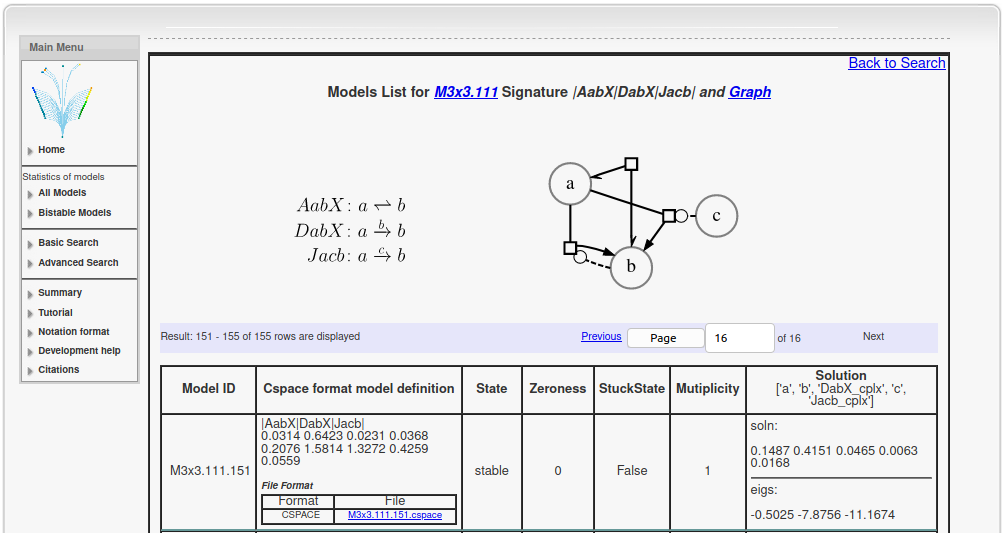
SBGN notation :
Above you can see an example page for the topology |AabX|DabX|Jacb|. On top of this page, one can see the set of chemical reactions in this topology on the left, with the corresponding chemical reaction network in the SBGN format on the right. In the SBGN notation, large circles represent reactant pools, squares with arrows represent directionality of the reaction and dotted lines with circles denote catalysis. Below this is a table with selected model parameters and solutions. Since this model has only one stable fixed point, it is monostable.
Model ID and the CSPACE format have been described above. For more details, please read the Development help page and the parent publication (Ramakrishnan and Bhalla, 2008).
State refers to the kind of fixed point found for the given model, analyzed using the eigenvalues of the Jacobian matix around the solution.Thus, the solution may be stable, unstable, saddle, line or doublet solution.
Zeroness specifies how many of the values in the solution vector are zero.
StuckState is "True" when the transition probability between stable states of the system in exactly zero.
Multiplicity specifies how many such identical solutions turned up.
----------------------------------------------------------------------
| soln: | 0.1487 | 0.4151 | 0.0465 | 0.0063 | 0.0168
|
Here we list the concentrations of the molecules at the solved steady-state.
The solution vector includes both the main set of molecules and also the enzyme-substrate complexes.
The order is as follows:
- Molecules are ordered alphabetically in case there are only "non-enzymatic reactions"
- In case there are only enzymatic or a mixture of non-enzymatic and enzymatic reactions, the position of an enzyme complex is after all the
- pure molecules taking part in the reaction, but before any later molecule.
- within this criterion, enzymes are ordered alphabetically.
- With this description,
- If you just had one reaction |CabX|, the order would be a, b, CabX_cplx,
- For the signature is |CabX|DacX|Fabc|. The solutions are in order: a, b, CabX_cplx, c, DacX_cplx.
- Similarly, for reactions involving three molecules, the enzyme-substrate complex would appear after all three pure molecules involved in the reaction.
- Thus for |Jabc|, the order will be a, b, c, Jabc_cplx.
- Taking another example, consider again the topology with the signature |AabX|DabX|Jacb|. The solutions for this topology will be in the order: a, b, DabX_cplx, c, Jacb_cplx.
----------------------------------------------------------------------
| eigs: | -0.5025 | -7.8756 | -11.1674 |
This lists the eigenvalues.
If all the eigenvalues are negative then solution will be a stable point.
If there is a single positive eigenvalue, it is a saddle.